Abstract
Background: Monoclonal gammopathy of undetermined significance (MGUS) is a benign hematological condition with the potential to progress to malignant conditions including multiple myeloma and Waldenstrom macroglobulinemia. To date, multiple clinical and laboratory findings have been identified as risk factors for MGUS progression, but any medications that modify progression risk have yet to be identified. To investigate this issue further, we propose a low-cost, easily replicated method for screening drug repurposing candidates in MGUS using real-world EHR data.
Methods: We extracted MGUS clinical and laboratory data from a manually curated MGUS database, containing 16,752 MGUS patients diagnosed from January 1, 2000, through December 31, 2021, prospectively maintained at Mayo Clinic. We then separately extracted medication data and comorbidities data, corresponding to the time from MGUS diagnosis to last follow-up or disease progression, from our electronic health record (EHR) database. Medications were mapped to 21 drug classes of interest and coded as the presence or absence of a drug from a given class, as there were challenges with ascertaining medication durations from our EHR data. The XGBoost gradient-boosted tree module was then used to fit a Cox survival model to the data, using a standard 70/15/15 train/validation/test split of the data, with 5-fold cross validation. The impact of explanatory features was quantified as hazard ratios using Shapley values averaged over 1000 repetitions of bootstrapping. For continuous features, hazard ratios were calculated by comparing the mean Shapley values for those above and below the median value of the continuous feature; for binary features, hazard ratios were calculated by comparing mean Shapley values for those with 0 versus 1 for a given feature.
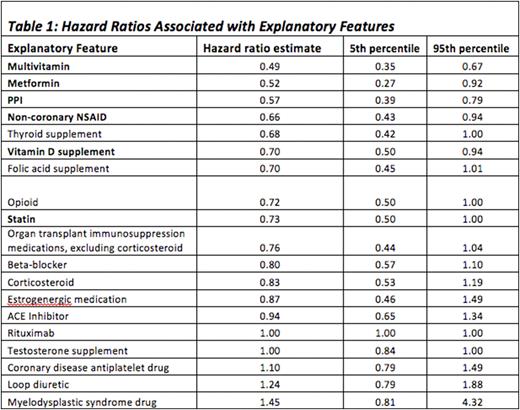
Results: Of the 16,752 patients in the MGUS database, medications data were available for 12,253; those without medications data were excluded. Our Cox survival model achieved a satisfactory fit of the data with inverse probability of censoring weights concordance index of 0.764. Within the medication groups examined, the presence of multivitamins, metformin, proton-pump inhibitors, non-coronary NSAIDs, vitamin D supplements or statins in patients’ medication lists were associated with significantly lower hazard ratio for progression. The full list of drug classes and associated hazard ratios are described in Table 1.
Discussion: We describe a low-cost method for using real-world EHR data and machine-learning to screen for drug repurposing candidates in MGUS. Our analysis identifies statins, metformin, multivitamins and opioids as having potential protective effects for MGUS progression. This work could inform subsequent prospective studies. Similar approaches can be replicated in other disorders allowing development of hypothesis driven research and repurposing of common drugs.
Disclosures
Kumar:AbbVie,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Adaptive,: Membership on an entity's Board of Directors or advisory committees, Research Funding; KITE,: Research Funding; MedImmune/Astra Zeneca,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Merck,: Research Funding; Novartis,: Research Funding; Roche: Research Funding; Sanofi: Research Funding; Oncopeptides: Other: Independent review committee. Dispenzieri:Oncopeptides, and Sorrento: Other: Data monitoring safety committee; Janssen: Membership on an entity's Board of Directors or advisory committees; Alynlam, Pfizer, Takeda, and BMS: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal